11 Introduction to R and RStudio
11.2 Learning outcomes
By the end of this tutorial you should be able to:
- Identify the different components of RStudio.
- Declare variables in R.
- Identify common data types and structures used in R.
- Recognize and use functions.
- Install and load R packages.
- Interpret documentation for functions and packages.
You should have both R and RStudio installed. Please open RStudio and work along the examples.
11.3 A Tour of RStudio
When you start RStudio, you will see something like the following window appear:
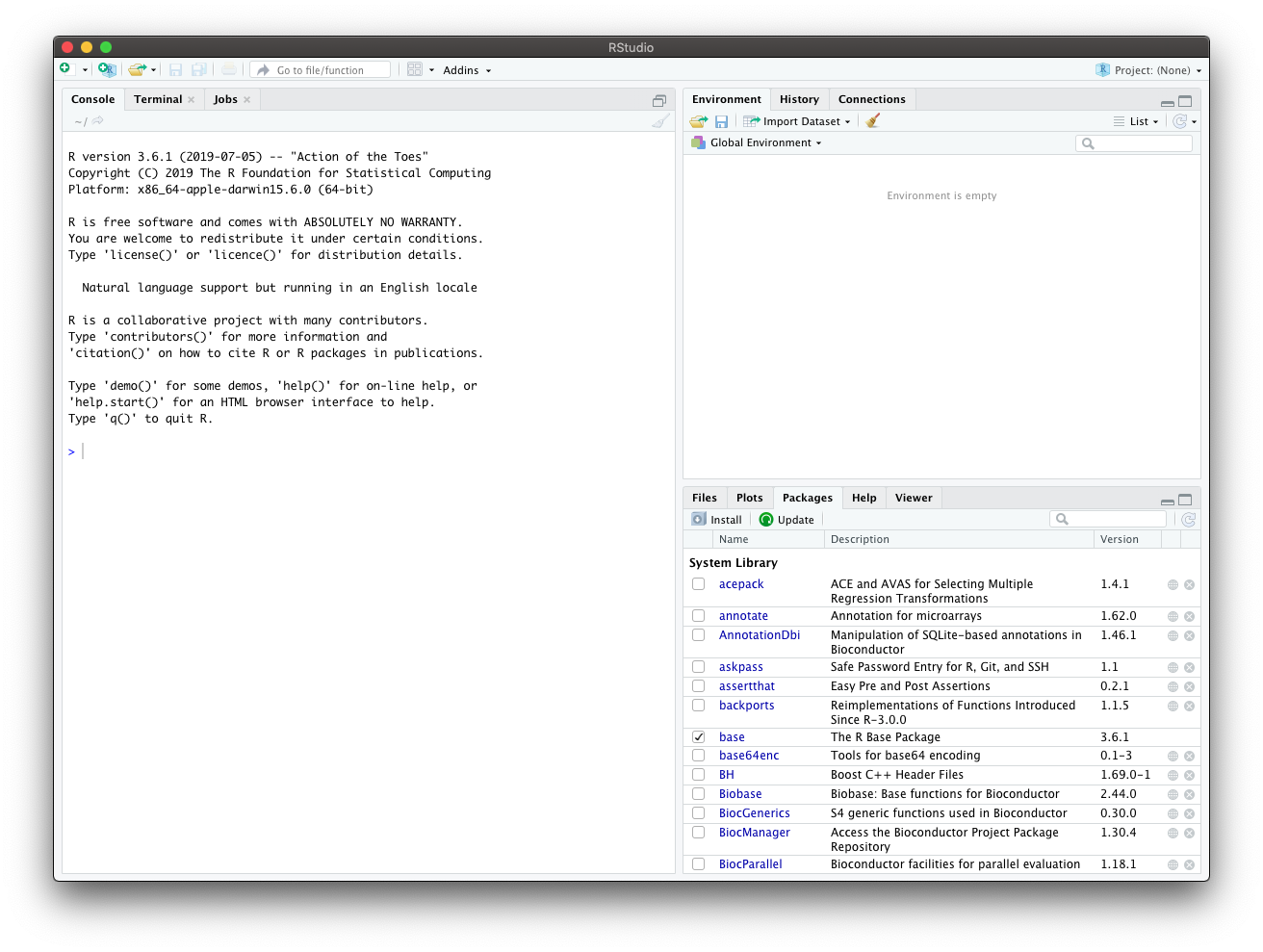
Notice that the window has three “panes”:
Console (lower left side): this is your view of the R engine. You can type in R commands here and see the output printed by R. (To tell them apart, your input is in blue, and the output is black.) There are several editing conveniences available: use up and down arrow keys to go back to previously entered commands, which you then can edit and re-run; TAB for completing the name before the cursor; see more in online docs.
Environment/History (tabbed in the upper right): view current user-defined objects and previously-entered commands, respectively.
Files/Help/Plots/Packages (tabbed in the lower right): as their names suggest, you can view the contents of the current directory, the built-in help pages, and the graphics you created, as well as manage R packages.
To change the look of RStudio, you can go to Tools > Global Options > Appearance and select colours, font size, etc. If you plan to be working for longer periods, we suggest choosing a dark background colour scheme to save your computer battery and your eyes.
11.4 Setup
11.4.1 RStudio Projects
Projects are a great feature of RStudio. When you create a project, RStudio creates an .Rproj file that links all of your files and outputs to the project directory. When you import data from a file, R automatically looks for it in the project directory instead of you having to specify a full file path on your computer. R also automatically saves any output to the project directory. Finally, projects allow you to save your R environment in .RData so that when you close RStudio and then re-open it, you can start right where you left off without re-importing any data or re-calculating any intermediate steps.
RStudio has a simple interface to create and switch between projects, accessed from the button in the top-right corner of the RStudio window. (Labeled “Project: (None)”, initially.)
Let’s create a project to work in for this tutorial. Start by choosing from the menu File > New Project. Select “New Project”, and the following will appear:
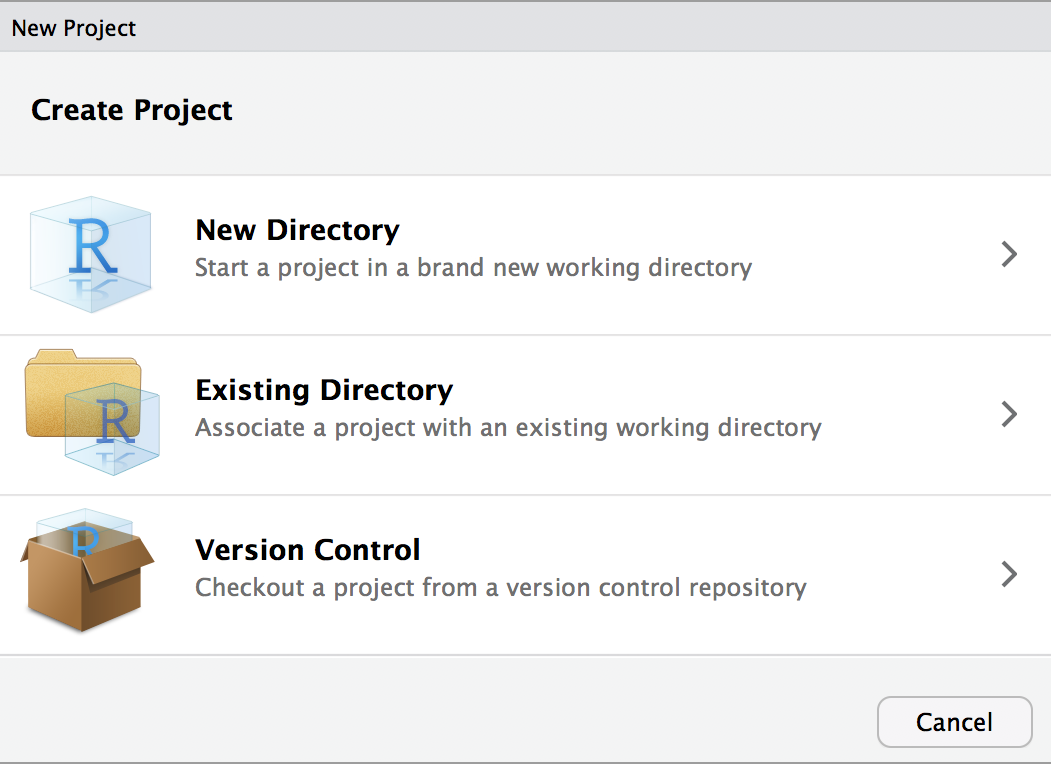
Choose “New Directory” followed by “New Project” and click on “Browse…”. Navigate to your Desktop, and name the directory deseq for this project.
After your project is created, navigate to its directory using your Finder (macOS) or File Explorer (Windows). You will see the deseq.Rproj file has been created.
You can open this project in the future in one of three ways:
- In your file browser (e.g. Finder or Explorer), simply double-click on the
deseq.Rprojfile. - In an open RStudio window, choose File > Open Project.
- Switch among projects by clicking on the R project symbol in the upper left corner of RStudio.
11.4.2 R packages
R packages are units of shareable code, containing functions that facilitate and enhance analyses. In simpler terms, think of R packages as iPhone Applications. Each App has specific capabilities that can be accessed when we install and then open the application. The same holds true for R packages. To use the functions contained in a specific R package, we first need to install the package, then each time we want to use the package we need to “open” the package by loading it.
While installing packages you might be prompted:
There is a binary version available but the source version is later:
<table of package names>
Do you want to install from sources the package which needs compilation? (Yes/no/cancel)Always enter no followed by enter.
In the following tutorials, we will be using the {tidyverse}, {pheatmap}, {BiocManager}, {DESeq2} and {plyranges} packages. {tidyverse} contains a versatile set of functions designed for easy manipulation of data. {pheatmap} visualizes gene expression levels and groups genes with similar patterns across treatments. {BiocManager} allows us to install packages from the Bioconductor platform (see below). Finally, {DESeq2} compares gene expression across treatments and {plyranges} provides a {tidyverse}-like interface for working with genomic ranges.
These packages are distributed on two different platforms, CRAN (Comprehensive R Archive Network) and Bioconductor (think of those platforms as two different app stores for R packages).
11.4.2.1 Installing Packages from CRAN
Packages from CRAN (Comprehensive R Archive Network) can be installed with the function install.packages().
install.packages(c("tidyverse", "pheatmap", "BiocManager"))11.4.2.2 Installing packages from Bioconductor
Bioconductor specializes on distributing R packages for the analysis of high-throughput genomic data. Unfortunately, install.packages() is limited to installing packages from CRAN only and cannot install any from Bioconductor. Luckily, we can download the package BiocManager from CRAN that then in turn provides an interface to access packages from Bioconductor.
This will be our first example of using a function provided by a package. In many situation, you just want to use a single function for a single time during your coding session. You can access the function by calling it with <package>::<function>(). As a concrete example, we can use the install() function of the BiocManager by using BiocManager::install() to install a Bioconductor package.
11.4.2.3 Loading packages
We sometimes will use functions of a package multiple times. To access the package’s functions and every time you open a new RStudio session, you need to first load (open) the packages you want to use with the library() function.
Packages can be loaded like this:
11.4.3 R Scripts
R script files are the primary way in which R facilitates reproducible research. They contain the code that loads your raw data, cleans it, performs the analyses, and creates and saves visualizations. R scripts maintain a record of everything that is done to the raw data to reach the final result. That way, it is very easy to write up and communicate your methods because you have a document listing the precise steps you used to conduct your analyses. This is one of R’s primary advantages compared to traditional tools like Excel, where it may be unclear how to reproduce the results.
Generally, if you are testing an operation (e.g. what would my data look like if I applied a log-transformation to it?), you should do it in the console (left pane of RStudio). If you are committing a step to your analysis (e.g. I want to apply a log-transformation to my data and then conduct the rest of my analyses on the log-transformed data), you should add it to your R script so that it is saved for future use.
Additionally, you should annotate your R scripts with comments. In each line of code, any text preceded by the # symbol will not execute. Comments can be useful to remind yourself and to tell other readers what a specific chunk of code does.
Let’s create an R script (File > New File > R Script) and save it as deseq.R in your deseq project directory. If you again look to the project directory on your computer, you will see deseq.R is now saved there.
We can copy and paste the previous library() commands in this tutorial and aggregate it in our R script.
11.5 Data types
When working with data in R, you might have noticed that not all data are the same, and these different forms of data are called data types. As part of this tutorial, you will encounter three basic data types:
| Data type | Description |
|---|---|
| character | Any type of text. Must be enclosed in quotes "<your text>"
|
| numeric | Decimal numbers |
| logical | Can only take TRUE or FALSE
|
Let’s look at a few examples of different data types. Below is a simple example of the type character. Character types are always enclosed by quotes.
salutation <- "Hello, World!"
salutation## [1] "Hello, World!"Here an example of a numeric:
some_number <- 6
some_number## [1] 6It is important to use the correct data type for a function or other operations in R. For example, the operator + only works with numeric data
6 + 2## [1] 8but not character data
"6" + "2"## Error in "6" + "2": non-numeric argument to binary operatorLogicals can take either TRUE or FALSE. You might have already encountered them as values for arguments in functions.
microbiology_is_awesome <- TRUE
microbiology_is_awesome## [1] TRUE11.6 Functions
Functions are one of the basic units in programming. Generally speaking, a function takes some input and generates some output. Every R function follows the same basic syntax, where function() is the name of the function and arguments are the different parameters you can specify (i.e. your input):
function(argument1 = ..., argument2 = ..., ...)
You can treat functions as a black box and do not necessarily need to know how it works under the hood as long as your provided input conforms to a specific shape.
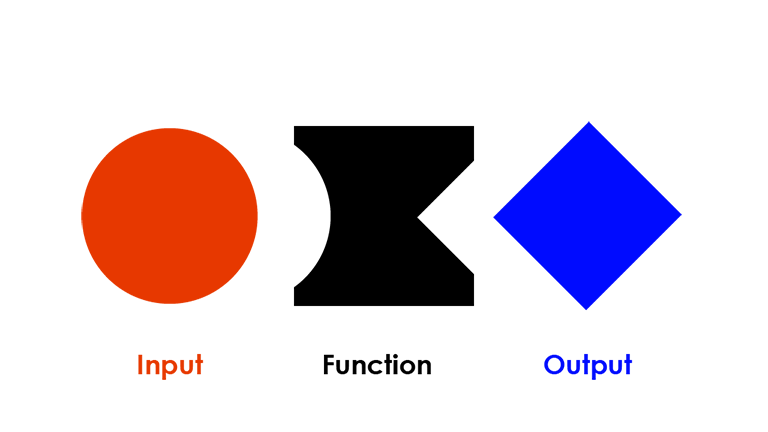
For example, the function sum() expects numbers:
sum(3, 5, 9, 18)## [1] 35If you instead pass text as arguments to sum() you will receive an error:
sum("Sum", "does", "not", "accept", "text!")## Error in sum("Sum", "does", "not", "accept", "text!"): invalid 'type' (character) of argumentSometimes, you want to confirm the data type (see above) of a variable, and you can do so by using the typeof() function.
typeof(microbiology_is_awesome)## [1] "logical"11.7 Getting Help
11.7.1 The help function
You can get help with any function in R by inputting ?function_name into the Console. If the function is part of a package, then you need to load the package first (i.e. with library()). This will open a window in the bottom right under the Help tab with information on that function, including input options and example code.
?read_delimThe Description section tells us that read_delim() is a general case of the function we used, read_csv(), and read_tsv().
The Usage section tells us the inputs that need to be specified and default inputs of read_delim:
-
fileanddelimneed to be specified by you as they do not have a default value (i.e. they are not followed by=). - All other parameters followed by
=and a value have a default (e.g.quote = "\") and do not have to be specified to run the function.
Reading technical documentation can be very confusing at first. It can be very helpful to first focus on the arguments of a function that have to be provided by you.
The Arguments Section describes the requirements of each input argument in detail.
The Examples Section has examples of the function that can be directly copy and pasted into your terminal and ran.
You can also get the description for a package itself, for example ?tidyverse. At the bottom you will find a link to an index that often will provide you with links to more documentation.
11.7.2 Vignettes
A lot of packages also provide a long-form guide called vignette to their functionality often giving examples of use-cases or more information on methods used. You can browse all vignettes with browseVignettes() or get the vignette for a particular package with browseVignettes("<package name>") (replace <package name> with name of your package).
Help for log() function
Using help to identify the necessary arguments for the log() function, compute the natural logarithm of 4, base 2 logarithm of 4, and base 4 logarithm of 4.
You have never used the function inner_join() of the dplyr package before. Take a look at the help documentation for inner_join() in RStudio (Hint: You can only look at the documentation for functions of packages you have loaded). In the list below, identify all of the arguments of the function that are mandatory and have to be specified by you.
xybycopysuffixkeep
What data type is accepted by the keep argument?
11.8 Data structures
The data types in R are the elements to build more complex objects called data structures. The most basic structures are vector, lists, matrix and data frame.
11.8.1 Vector
We can use the c() function to create a vector. All vector elements have to be of the same data type (a so-called atomic vector).
vector_example <- c("This", "vector", "has", "5", "elements!")
vector_example## [1] "This" "vector" "has" "5" "elements!"Coercion
Since all vector elements have to be of the same type, what happens if we use c() with dissimilar ones? To test R’s behaviour, create several vectors of all possible pairwise combinations of character, numeric (use any integer), and logical, then determine the type of the object using typeof(<your vector>). In addition, test the combination of (a) logical with either the numbers 0 or 1 or (b) logical and an integer larger than 1 or smaller than -1.
The behaviour you will observe is called coercion. Why do you think this particular set of behaviours were selected for the design of R?
You can also assign each element a name using <name> = <value>
vector_example <- c(first = "Hi", second = "there")
vector_example## first second
## "Hi" "there"11.8.2 List
In R, a list is a special kind of vector whose elements can be any combination of objects (i.e. they also do not have to be the same).
You can also assign a name to elements of a list.
11.8.3 Matrix
The data is provided as a vector to matrix() and filled either column- or row-wise into the matrix. Rows and columns can also be assigned names.
matrix_example <- matrix(
data = c(
10, 20,
3, 6
),
byrow = TRUE,
ncol = 2,
dimnames = list(
c("gene_a", "gene_b"),
c("control", "treated")
)
)
matrix_example## control treated
## gene_a 10 20
## gene_b 3 611.8.4 Data frame
A data frame is another data structure that contains elements in columns and rows and it is a list of vectors, with each column being one vector of a single data type but the data type may differ among vectors. Data frames are a big part of working with data in R. Data is always provided by column <column name> = <data>
data_frame_example <- data.frame(
gene_name = c("gene_a", "gene_b"),
control = c(10, 3),
treated = c(20, 3)
)
data_frame_example## gene_name control treated
## 1 gene_a 10 20
## 2 gene_b 3 3Although the data frame and matrix conain the same information, they are not the same class of object. You can determine the class with class().
class(matrix_example)## [1] "matrix" "array"
class(data_frame_example)## [1] "data.frame"11.8.5 Subsetting
We often want to use only part of the data and have to subset them accordingly. In base R, subsetting is done with brackets [ ]. For example, a vector can be subset by the index or the name of the position.
vector_example[1]## first
## "Hi"
vector_example["first"]## first
## "Hi"Matrices and data frames can also be subset that way by first specifying rows then columns [<rows>, <columns>].
matrix_example[, "control"]## gene_a gene_b
## 10 3
matrix_example[1, "control"]## [1] 10We can extract a single column (vector) from a data frame using the $ operator.
data_frame_example[, "control"]## [1] 10 3
data_frame_example$control## [1] 10 3