2 A tutorial for using TreeSAPP
2.1 Introduction and goals
In this series of tutorials, students will analyze a gene family by creating a TreeSAPP reference package (refpkg). Students will work through a typical workflow of TreeSAPP both with an example gene (XmoA) to familiarize yourself with the tools, and then you will repeat the steps with a gene assigned to your group for which no reference package exists. You will document your efforts for the new reference package in Problem Set 5.
2.2 TreeSAPP workflow
TreeSAPP is a Python package for gene-centric analysis. It uses custom protein sequence databases called reference packages (RefPkg)
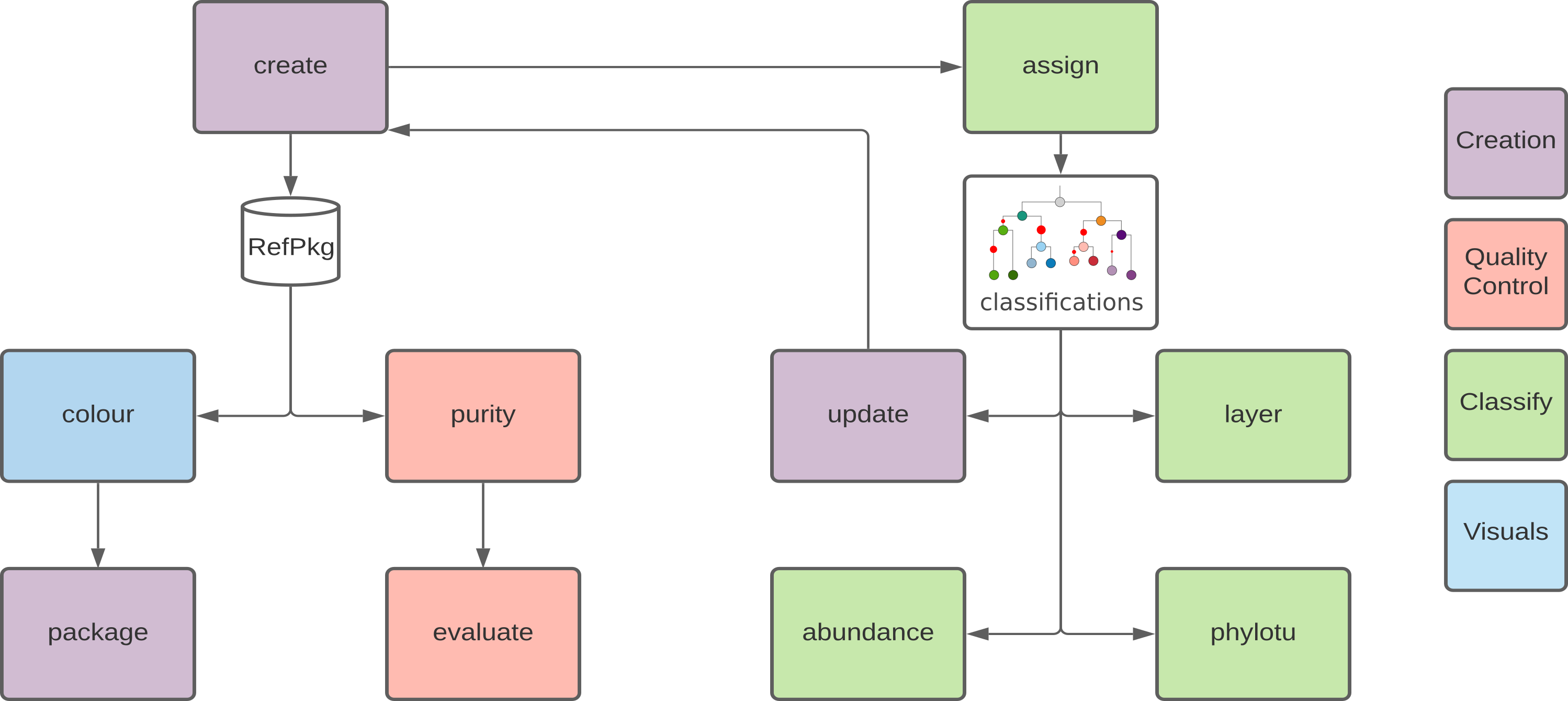
2.3 Genes for creating reference packages
Reference packages can be built for nearly any protein-encoding gene but to demonstrate the process of gene-centric analysis we will create and use a reference package for XmoA.
2.3.1 XmoA
The protein family we will be focusing on is that of the copper-containing membrane-bound monooxygenases (5). This family contains particulate methane monooxygenase (pMMO) and ammonia monooxygenase (AMO) and well be building a reference package for the alpha subunits of these enzymes called XmoA. All students will work through this example individually.
2.4 Tools
2.4.1 Shell
Please use this short Shell cheat sheet for commonly used commands and review previous tutorials on Canvas.
2.5 Data
Other than The Saanich Inlet data set already located on the server, you may download data from different databases: